For large-scale data, you can also try the local version: https://github.com/KennthShang/PhaBOX
Introduction
⌛️ News
PhaBOX has now been upgraded to the 2.0 version with faster speed!
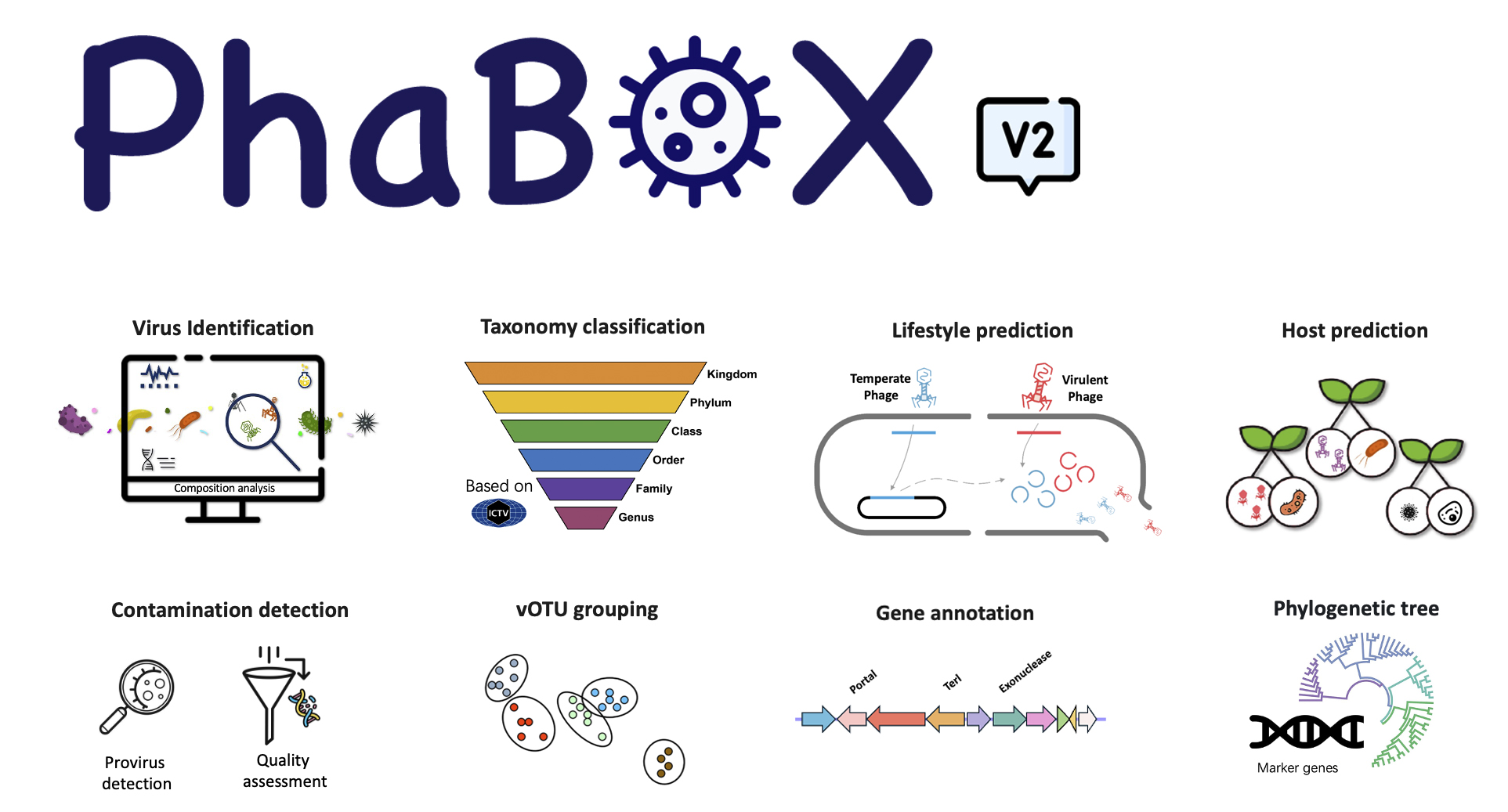
There are some major components, including:
- 🎉 Generalized for all kinds of viruses
- 🎉 Virus identification
- 🎉 Taxonomy classification
- 🎉 Host prediction
- 🎉 Lifestyle prediction
- 🎉 Contamination/provirus detection
- 🎉 vOTU grouping
- 🎉 Phylogenetic tree based on marker genes
- 🎉 Viral protein annotation
- 🎉 All the databases are updated to the latest ICTV 2024 release
If you have any more suggestions, feel free to let me know! We consider long-term maintenance PhaBOX and adding modules according to your needs. You can post an issue or directly email me (jiayushang@cuhk.edu.hk). We welcome any suggestions.
🚀 Quick Start
Please check our Tutorial page. We provide a tutorial to help you get started quickly and understand how to use PhaBOX2. We hope you will enjoy it!
An example of the standard output can be found via EXAMPLE.
💻 Local version
This is the source code of our website PhaBOX2.
📘 License
The PhaBOX pipelines are released under the terms of the Academic Free License v3.0 License.
